Deep learning instance segmentation
Deep learning based nuclei and cell segmentation can be much faster and more accurate than conventional segmentation methods. In addition, there may be less parameter tuning required than for conventional methods. However, one can still make mistakes, such as applying the wrong deep learning model for the given input data. Both the usefulness and potential pitfalls make it very important to learn how to properly use and judge deep learning based image segmentation.
Prerequisites
Before starting this lesson, you should be familiar with:
Learning Objectives
After completing this lesson, learners should be able to:
Run a deep learning model for instance segmentation
Visually assess the quality of the segmentation
Appreciate that, in order to work well, the model has to match the input data
Appreciate that even deep learning models may have parameters that need to be tuned
Appreciate that subtle differences in the image data may cause the model to fail
Concept map
Figure
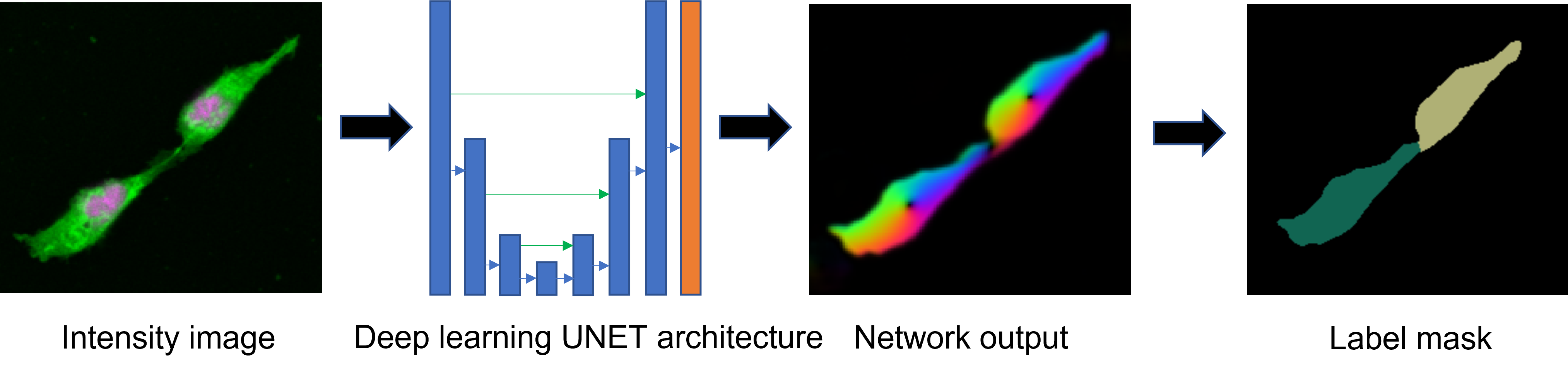
Activities
- Use a deep learning tool to segment the cells in this image: xyc_8bit__membranes_nuclei.tif.
- If the tool allows you to run different models, try them and compare the results
- If the tool has parameters, explore them and compare the results
Show activity for:
CellPose GUI
- Download this two-channel image.
- Open the image in Fiji and measure the typical diameter of one cell.
- Open the image in Cellpose
cell diameter: 60chan to segment: red(membranes)chan2 (optional): green(nuclei)- Run the
cytomodel
- Appreciate that the cell segmentation works well
- Decrease the
cell diameterto 1/5 of its correct value- Run the
cytomodel again
- Appreciate that the segmentation does not work well
- Put the
cell diameterback to60- Run the
nucleimodel
- Appreciate that also this does not work well
- Appreciate that only the correct parameters and model yield a good result
Debug a tricky case (optional)
- Download a similar two-channel image.
- Try to segment it in CellPose
- Appreciate that some cells are not segmented
- Try to find out why that could be the case
Explore additional parameters (optional)
Explore the
flow_threshold,cell_thresholdandstitch_thresholdparameters
Exercises
Show exercise/solution for:CellPose GUI
- Download this two-channel image.
- Open the image in
Cellpose
cell diameter: 60chan to segment: 1 red(membranes)chan2 (optional): 0 none- Run the
cytomodelDo you have a different opinion on the segmentation, if yes, can you locate regions where you think
cellposemay have segmented differently?Solution
Hint: Use
MASKS ON [x]andoutlines on [Z]to view the problematic areas in much intuitive way. Use arrow keys up and down to switch between channels and different display modes. Increase contrast if necessary. The problematic segmented areas are shown here:Play with
Cell diameter,chan to segmentandchan2(optional)and find parameter values to improve the segmentation from the preceding stepSolution
One way to do this is to use nuclear channel information by setting
chan2 (optional): 2 greenand runcytomodel again. Another way to make results better is to use a lowerCell diameter = 40
Assessment
Fill in the blanks
- The typical output of a (deep learning based) instance segmentation is a ___ .
- If the segmentation is faulty you could consider to __ or __ .
- If you wonder which deep learning tool to use to segment your data you could _ or __.
- To run a deep learning model efficiently you should have a computer with ___ .
Solution
- label mask image
- use a different model or change the tool’s parameters
- ask on the forum or check the BioImage Model Zoo
- a GPU
Follow-up material
Recommended follow-up modules:
Train a model (TODO)
Learn more:
