Output saving
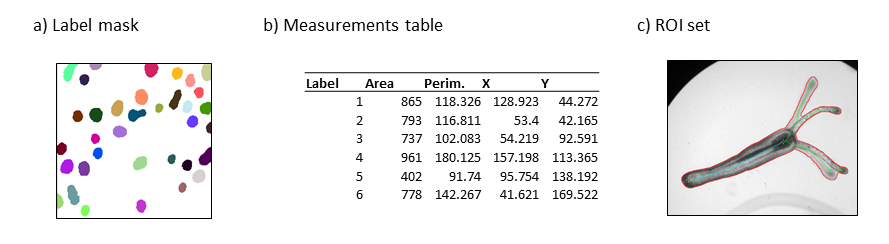
There are multiple situations in which you need to save the different types of output you can generate with your image analysis pipelines. For example, you may want to save your results as measurement tables for further analysis in other software (e.g. RStudio, MS EXCEL, ..). It can also be important to save the regions of interest (ROIs) that were used for particular measurements, so that you can look back at them for reference, or to use them for visualization purposes. In addition to ROIs, images can be saved as label masks to store the information on different regions. Finally, you may also want to save your entire script containing particular settings or parameters that you used, so that you can re-run the analysis with the exact same settings or compare it with the results obtained using different settings.
Prerequisites
Before starting this lesson, you should be familiar with:
Generating results tables
Defining regions of interest (ROIs)
Learning Objectives
After completing this lesson, learners should be able to:
Save measurements as a table
Save ROIs
Save output label mask
Concept map
Figure
Activities
- Use a template script and run it.
- Understand what the commands do.
- Record yourself saving the output.
- Edit the script using the recorded commands.
- Optional: define an output directory as a parameter and use string concatenation to specify where to save the file and with what name. An example output directory could be ‘C:\\Users\\username\\Desktop’ on Windows (note double “\” because a single “\” is interpreted as an escape character) or ‘/Users/username/Desktop/’ on MacOS.
Show activity for:
ImageJ Macro
- Download the template script: output_saving_imagej-macro_template.ijm. The aim of the script is to generate different kinds of output (labels, results, ROIs) from this image: xy_8bit_binary_randomshapes.tif.
- Run and understand the script.
- Add commands that save the different output: label image, results table, ROIs.
Solution
// This macro uses the particle analyzer to measure features of shapes. // Different outputs are saved: ROIs, results table, and label mask. // make sure the background is set to black in Process>Binary>Options run("Options...", "iterations=1 count=1 black do=Nothing"); // specify an output directory outputDir = FIXME // (e.g. 'C:\\Users\\username\\Desktop' or 'C:/Users/username/Desktop' on Windows, or '/Users/username/Desktop/' on MacOS) // specify size parameters for object selection minSize = 0 maxSize = 1000 // close any pre-existing output you do not want in your saving results roiManager("reset"); // clear any pre-existing ROIs run("Clear Results"); // clear any pre-existing results run("Close All"); // close any open images // Set measurements and run particle analyzer on binary shapes image run("Set Measurements...", "area centroid center perimeter bounding redirect=None decimal=3") // set desired measurements open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary_randomshapes.tif"); // open binary image with random shapes run("Analyze Particles...", "size=&minSize-&maxSize show=[Count Masks] display add") // run the particle analyzer>run("glasbey") // Save the results saveAs("tiff", outputDir + File.separator + "shapes_labels_macro.tiff"); // save label mask to output directory saveAs("results", outputDir + File.separator + "shapes_results_macro.txt"); // save results file to output directory roiManager("Save", outputDir + File.separator + "shapes_ROIset_macro.zip"); // save rois to output directory
ImageJ Jython
- Download the template script: output_saving_imagej-jython_template.py. The aim of the script is to generate different kinds of output (labels, results, ROIs) from this image: xy_8bit_binary_randomshapes.tif.
- Run and understand the script.
- Add commands that save the different output: label image, results table, ROIs.
Solution
# import classes from ij import IJ, ImagePlus, WindowManager from ij.io import FileSaver from ij.plugin.filter import ParticleAnalyzer from ij.plugin.frame import RoiManager from ij.measure import ResultsTable, Measurements from ij.process import ImageProcessor import os # make sure the background is set to black in Process>Binary>Options IJ.run("Options...", "iterations=1 count=1 black") # Specify an output directory outputDir = FIXME # (e.g. r'C:\Users\username\Desktop', 'C:\\Users\\username\\Desktop' or 'C:/Users/username/Desktop' on Windows or '/Users/username/Desktop/' on MacOS) # Specify size parameters for object selection min_size = 0 max_size = 1000 # Initialize Roi Manager and empty results table, close other open images rm = RoiManager().getInstance() rm.reset() IJ.run("Close All") # Open binary shapes image shapes = IJ.openImage("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary_randomshapes.tif") # Configure and run particle analyzer results = ResultsTable() # construct empty resultstable pa = ParticleAnalyzer((ParticleAnalyzer.ADD_TO_MANAGER + ParticleAnalyzer.SHOW_ROI_MASKS),(Measurements.AREA + Measurements.CENTROID + Measurements.CENTER_OF_MASS + Measurements.PERIMETER + Measurements.RECT), results, min_size, max_size, 0, 1) pa.analyze(shapes) # run the particle analyzer on the image results.show("Results") # Save label mask, results table, and ROIs labelMask = WindowManager.getImage("Count Masks of xy_8bit_binary_randomshapes.tif") IJ.run(labelMask, "Glasbey", "") # set glasbey LUT FileSaver(labelMask).saveAsTiff(os.path.join(outputDir, "shapes_labels_jython.tif")) # save the label mask results.save(os.path.join(outputDir, "shapes_results_jython.txt")) # save results table rm.runCommand("Save", os.path.join(outputDir, "shapes_ROIset_jython.zip")) # save the ROIs
Exercises
Show exercise/solution for:ImageJ Macro
Adapt the code from the activity such that you:
- Specify an output directory in the beginning
- Save the results table as comma-separated data table instead of text-delimited data.
- Save the output label image in a different image format (e.g. PNG, JPEG). Is this a good format for label images?
Solution
// This macro uses the particle analyzer to measure features of shapes. // Different outputs are saved: ROIs, results table, and label mask. // make sure the background is set to black in Process>Binary>Options run("Options...", "iterations=1 count=1 black do=Nothing"); // specify an output directory outputDir = FIXME // (e.g. 'C:\\Users\\username\\Desktop' or 'C:/Users/username/Desktop' on Windows, or '/Users/username/Desktop/' on MacOS) // specify size parameters for object selection minSize = 0 maxSize = 1000 // close any pre-existing output you do not want in your saving results roiManager("reset"); // clear any pre-existing ROIs run("Clear Results"); // clear any pre-existing results run("Close All"); // close any open images // Set measurements and run particle analyzer on binary shapes image run("Set Measurements...", "area centroid center perimeter bounding redirect=None decimal=3") // set desired measurements open("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary_randomshapes.tif"); // open binary image with random shapes run("Analyze Particles...", "size=&minSize-&maxSize show=[Count Masks] display add") // run the particle analyzer>run("glasbey") // Save the results saveAs("Png", outputDir + File.separator + "/shapes_labels_macro.png"); // save label mask to output directory saveAs("Results", outputDir + File.separator + "/shapes_results_macro.csv"); // save results file to output directory roiManager("Save", outputDir + File.separator + "/shapes_ROIset_macro.zip"); // save ROIs to output directory
ImageJ Jython
Adapt the code from the activity such that you:
- Specify an output directory in the beginning
- Save the results table as comma-separated data table instead of text-delimited data.
- Save the output label image in a different image format (e.g. PNG, JPEG). Is this a good format for label images?
Solution
# import classes from ij import IJ, ImagePlus, WindowManager from ij.io import FileSaver from ij.plugin.filter import ParticleAnalyzer from ij.plugin.frame import RoiManager from ij.measure import ResultsTable, Measurements from ij.process import ImageProcessor import os # make sure the background is set to black in Process>Binary>Options IJ.run("Options...", "iterations=1 count=1 black") # Specify an output directory outputDir = FIXME # (e.g. r'C:\Users\username\Desktop', 'C:\\Users\\username\\Desktop' or 'C:/Users/username/Desktop' on Windows or '/Users/username/Desktop/' on MacOS) # Specify size parameters for object selection min_size = 0 max_size = 1000 # Initialize Roi Manager and empty results table, close other open images rm = RoiManager().getInstance() rm.reset() IJ.run("Close All") # Open binary shapes image shapes = IJ.openImage("https://github.com/NEUBIAS/training-resources/raw/master/image_data/xy_8bit_binary_randomshapes.tif") # Configure and run particle analyzer results = ResultsTable() # construct empty resultstable pa = ParticleAnalyzer((ParticleAnalyzer.ADD_TO_MANAGER + ParticleAnalyzer.SHOW_ROI_MASKS),(Measurements.AREA + Measurements.CENTROID + Measurements.CENTER_OF_MASS + Measurements.PERIMETER + Measurements.RECT), results, min_size, max_size, 0, 1) pa.analyze(shapes) # run the particle analyzer on the image results.show("Results") # Save results, label mask, and ROIs labelMask = WindowManager.getImage("Count Masks of xy_8bit_binary_randomshapes.tif") IJ.run(labelMask, "Glasbey", "") # set glasbey LUT FileSaver(labelMask).saveAsPng(os.path.join(outputDir, "shapes_labels_jython.png")) # save the label mask results.save(os.path.join(outputDir, "shapes_results_jython.csv")) # save results table rm.runCommand("Save", os.path.join(outputDir, "shapes_ROIset_jython.zip")) # save the ROIs
Assessment
Fill in the blanks
- Three commonly saved data output types include ___ .
- In order to safely re-open your data or open it in different software, you need to save in an __ file format.
Solution
- Three commonly saved data output types include results tables, ROI sets, and label masks.
- In order to safely re-open your data or open it in different software, you need to save in an interoperable file format.
True or False
- Label masks should be saved in JPEG format.
- Tab-delimited text is a decent output format for results tables.
Solution
- Label masks should be saved in JPEG format. False (JPEG compression results in loss of the unique label values in the image)
- Tab-delimited text is a decent output format for results tables. True (this is generally more stable in other software than for example comma-delimited data)
Follow-up material
Recommended follow-up modules:
Learn more: