Image data formats course overview
Performing modern microscopy one is exposed to many different image data formats. Almost all microscope vendors have their own format. In addition, there are a few formats that are developed by the scientific community. Navigation this sea of formats is very challenging. In this course you will learn how to read most image formats and how to write a few recommened formats. In addition, you will learn about reading and writing big image data, which includes learning about the concepts of chunking and scale pyramids.
Prerequisites
Before starting this lesson, you should be familiar with:
Familiarity with using Fiji’s graphical user interface
Connected components labeling(label mask from segmentation)
Learning Objectives
After completing this lesson, learners should be able to:
Know and open various common image files formats
Understand important concepts and implementations of big image file formats: lazy-loading, chunking and scale pyramids
Open and create OME-TIFF
Open and create OME-Zarr
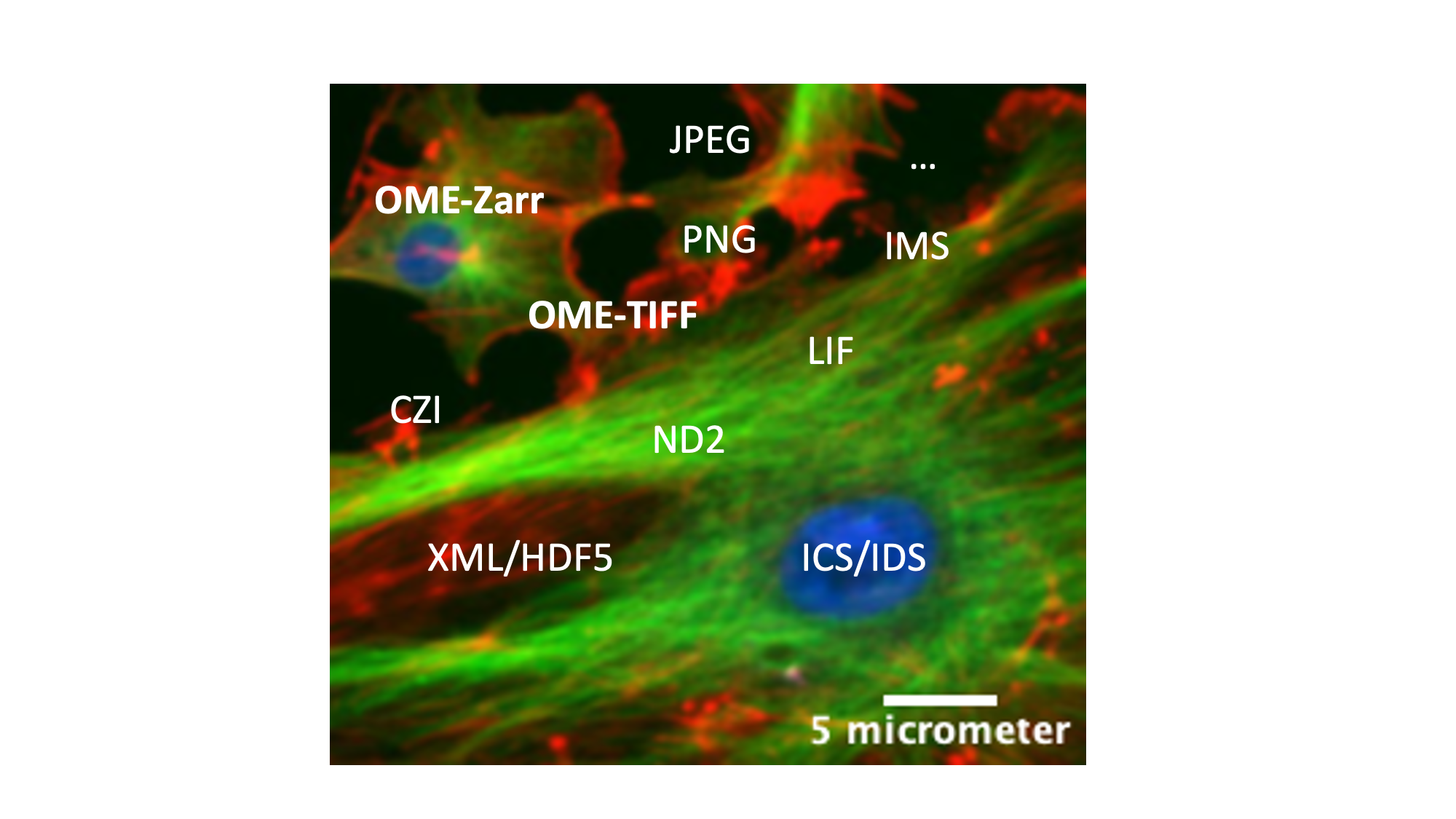
Concept map
Figure
General comments
The material for this course consists of the following training modules:
As usual, each training module contains several activities that can be executed in various compute platforms. As a trainer, it is necessary to familiarise yourself with the material and decide what to teach.
It is also highly recommended to consider our TEACHING TIPS.
Installation instructions
In order to conduct the activities it is required to perform the below installations before the course.
Install the python and command line environment
- Install the Miniforge3 conda package manager
- Create the environment
conda env create -f https://raw.githubusercontent.com/NEUBIAS/training-resources/refs/heads/master/_includes/tool_installation/image_data_formats_conda_env.yml - Activate the environment:
conda activate image_data_formats - Test the environment:
napari- The napari image viewer should open; if this fails, check the below troubleshooting section or contact the instructors
Conda installation troubleshooting
- Remove the environment:
conda env remove -n image_data_formats - Delete any caches:
conda clean -all - Also remove cached packages:
rm -r ~/miniforge3/pkgs - Then try installing the environment again following the instructions above
How to run this course
The current material allows to teach this course using Fiji and then command line interface, or using python. In our experience it works well to teach this course in 2 days. One day one, go through all modules using Fiji’s graphical user interface, on day two repeat the material using python. Day two could be optional, only for people that are interested in python scripting.
Previous editions of this course
For more details on how you may teach this course please check the detailed schedule of previous courses:
Follow-up material
Recommended follow-up modules:
Learn more: